
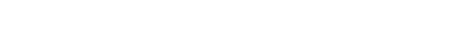
钍基核裂变能全国重点实验室 |
|||
|
-钍基核能物理中心-
|
|||
熔盐机械工程技术部 |
|||
仪控工程技术部 |
|||
熔盐化学工程技术部 |
|||
|
-核能综合利用研究中心-
|
|||
材料研究部 |
|||
钍铀循环化学部 |
|||
应用化学技术部 |
|||
氚科学与工程技术部 |
|||
核与辐射安全技术部 |
|||
应用加速器技术部 |
|||
反应堆运行技术部 |



报告名称:Investigating conformational changes of biological macromolecules using Markov State Models
报 告 人:Dr. Xuhui Huang
报告时间:2012年9月6日(星期四)上午10:00
报告地点:嘉定园区学术活动中心三楼报告厅
报告简介:
Simulating biologically relevant timescales at atomic resolution is a challenging task since typical atomistic simulations are at least two orders of magnitude shorter. Markov State Models (MSMs) built from molecular dynamics (MD) simulations provide one means of overcoming this gap without sacrificing atomic resolution by extracting long time dynamics from short MD simulations. MSMs coarse grain space by dividing conformational space into long-lived, or metastable, states. This is equivalent to coarse graining time by integrating out fast motions within metastable states. By varying the degree of coarse graining one can vary the resolution of an MSM; therefore, MSMs are inherently multi-resolution. In this talk, I will demonstrate the power of MSMs by applying it to simulate the complex conformational changes that occurs at tens of microsecond timescales for a large RNA transcription complex (close to half million atoms). In the second part of my talk, I will introduce a new efficient dynamic clustering algorithm for the automatic construction of MSMs for multi-body systems. We have successfully applied this new algorithm to model the protein-ligand recognition and hydrophobic collapse processes that occur at a mixture of different timescales.
报告人简介:
EDUCATION
07/2006, Ph.D in Chemical Physics, Chemistry Dept, Columbia University
06/2001, Bachelor of Science, University of Science and Technology of China
ACADEMIC POSITIONS
01/2010-Present, Assistant Professor, Chemistry Dept, the Hong Kong University of Science and Technology
09/2008-12/2009, Research Associate, Bioengineering Dept, Stanford University
08/2006-09/2008, Postdoctoral Scholar, Bioengineering Dept, Stanford University
ACADEMIC AWARDS
CCG Excellence Award, American Chemical Society, COMP Division, 2006
RESEARCH INTEREST
My recent research has been focused on understanding thermodynamics and kinetics of the protein folding and ligand binding using molecular simulations. I have developed various novel computational algorithms to enhance conformational sampling.